SomatiCA: identifying, characterizing, and quantifying somatic copy number aberrations from cancer genome sequencing
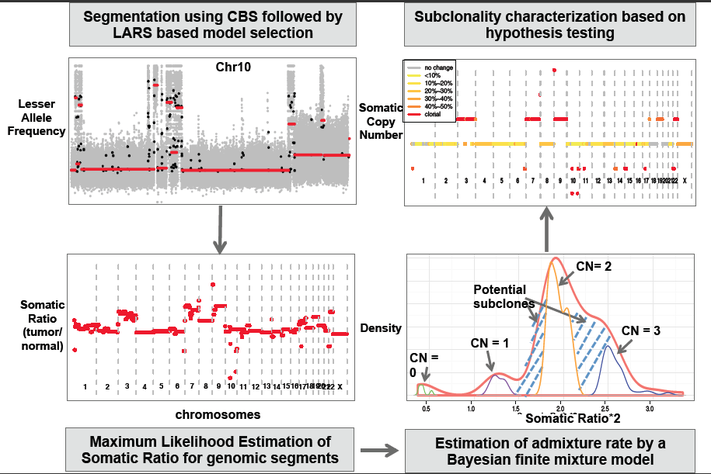
Whole genome sequencing of matched tumor-normal sample pairs is becoming routine in cancer research. However, analysis of somatic copy-number changes from sequencing data is still challenging because of insufficient sequencing coverage, unknown tumor sample purity and subclonal heterogeneity. Here we describe a computational framework, named SomatiCA, which explicitly accounts for tumor purity and subclonality in the analysis of somatic copy-number profiles. Taking read depths (RD) and lesser allele frequencies (LAF) as input, SomatiCA will output 1) admixture rate for each tumor sample, 2) somatic allelic copy-number for each genomic segment, 3) fraction of tumor cells with subclonal change in each somatic copy number aberration (SCNA), and 4) a list of substantial genomic aberration events including gain, loss and LOH.